structure(list(mpg = c(21, 21, 22.8, 21.4, 18.7, 18.1, 14.3,
24.4, 22.8, 19.2, 17.8, 16.4, 17.3, 15.2, 10.4, 10.4, 14.7, 32.4,
30.4, 33.9, 21.5, 15.5, 15.2, 13.3, 19.2, 27.3, 26, 30.4, 15.8,
19.7, 15, 21.4), cyl = c(6, 6, 4, 6, 8, 6, 8, 4, 4, 6, 6, 8,
8, 8, 8, 8, 8, 4, 4, 4, 4, 8, 8, 8, 8, 4, 4, 4, 8, 6, 8, 4),
disp = c(160, 160, 108, 258, 360, 225, 360, 146.7, 140.8,
167.6, 167.6, 275.8, 275.8, 275.8, 472, 460, 440, 78.7, 75.7,
71.1, 120.1, 318, 304, 350, 400, 79, 120.3, 95.1, 351, 145,
301, 121), hp = c(110, 110, 93, 110, 175, 105, 245, 62, 95,
123, 123, 180, 180, 180, 205, 215, 230, 66, 52, 65, 97, 150,
150, 245, 175, 66, 91, 113, 264, 175, 335, 109), drat = c(3.9,
3.9, 3.85, 3.08, 3.15, 2.76, 3.21, 3.69, 3.92, 3.92, 3.92,
3.07, 3.07, 3.07, 2.93, 3, 3.23, 4.08, 4.93, 4.22, 3.7, 2.76,
3.15, 3.73, 3.08, 4.08, 4.43, 3.77, 4.22, 3.62, 3.54, 4.11
), wt = c(2.62, 2.875, 2.32, 3.215, 3.44, 3.46, 3.57, 3.19,
3.15, 3.44, 3.44, 4.07, 3.73, 3.78, 5.25, 5.424, 5.345, 2.2,
1.615, 1.835, 2.465, 3.52, 3.435, 3.84, 3.845, 1.935, 2.14,
1.513, 3.17, 2.77, 3.57, 2.78), qsec = c(16.46, 17.02, 18.61,
19.44, 17.02, 20.22, 15.84, 20, 22.9, 18.3, 18.9, 17.4, 17.6,
18, 17.98, 17.82, 17.42, 19.47, 18.52, 19.9, 20.01, 16.87,
17.3, 15.41, 17.05, 18.9, 16.7, 16.9, 14.5, 15.5, 14.6, 18.6
), vs = c(0, 0, 1, 1, 0, 1, 0, 1, 1, 1, 1, 0, 0, 0, 0, 0,
0, 1, 1, 1, 1, 0, 0, 0, 0, 1, 0, 1, 0, 0, 0, 1), am = c(1,
1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1,
0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1), gear = c(4, 4, 4, 3,
3, 3, 3, 4, 4, 4, 4, 3, 3, 3, 3, 3, 3, 4, 4, 4, 3, 3, 3,
3, 3, 4, 5, 5, 5, 5, 5, 4), carb = c(4, 4, 1, 1, 2, 1, 4,
2, 2, 4, 4, 3, 3, 3, 4, 4, 4, 1, 2, 1, 1, 2, 2, 4, 2, 1,
2, 2, 4, 6, 8, 2)), row.names = c("Mazda RX4", "Mazda RX4 Wag",
"Datsun 710", "Hornet 4 Drive", "Hornet Sportabout", "Valiant",
"Duster 360", "Merc 240D", "Merc 230", "Merc 280", "Merc 280C",
"Merc 450SE", "Merc 450SL", "Merc 450SLC", "Cadillac Fleetwood",
"Lincoln Continental", "Chrysler Imperial", "Fiat 128", "Honda Civic",
"Toyota Corolla", "Toyota Corona", "Dodge Challenger", "AMC Javelin",
"Camaro Z28", "Pontiac Firebird", "Fiat X1-9", "Porsche 914-2",
"Lotus Europa", "Ford Pantera L", "Ferrari Dino", "Maserati Bora",
"Volvo 142E"), class = "data.frame")
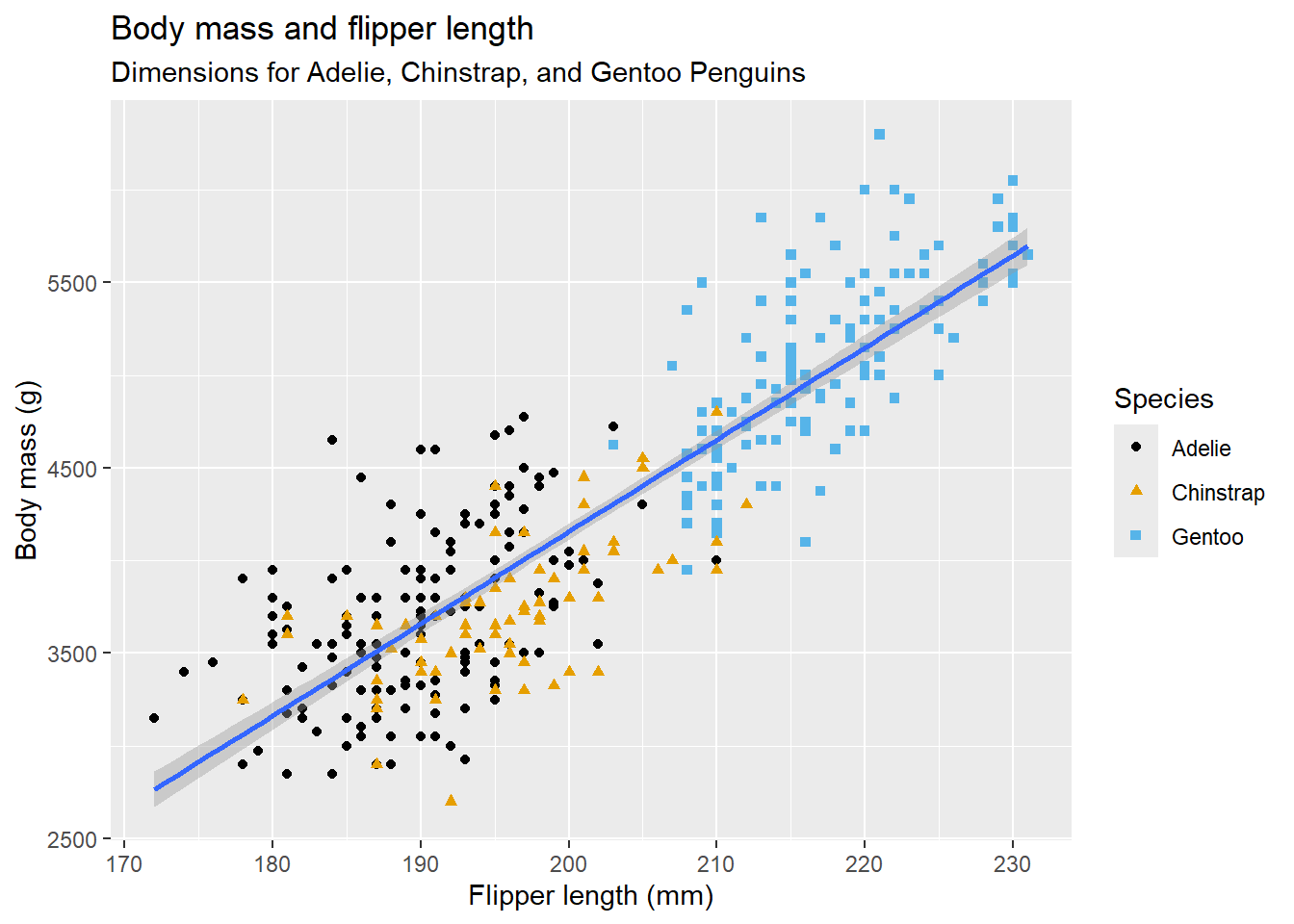
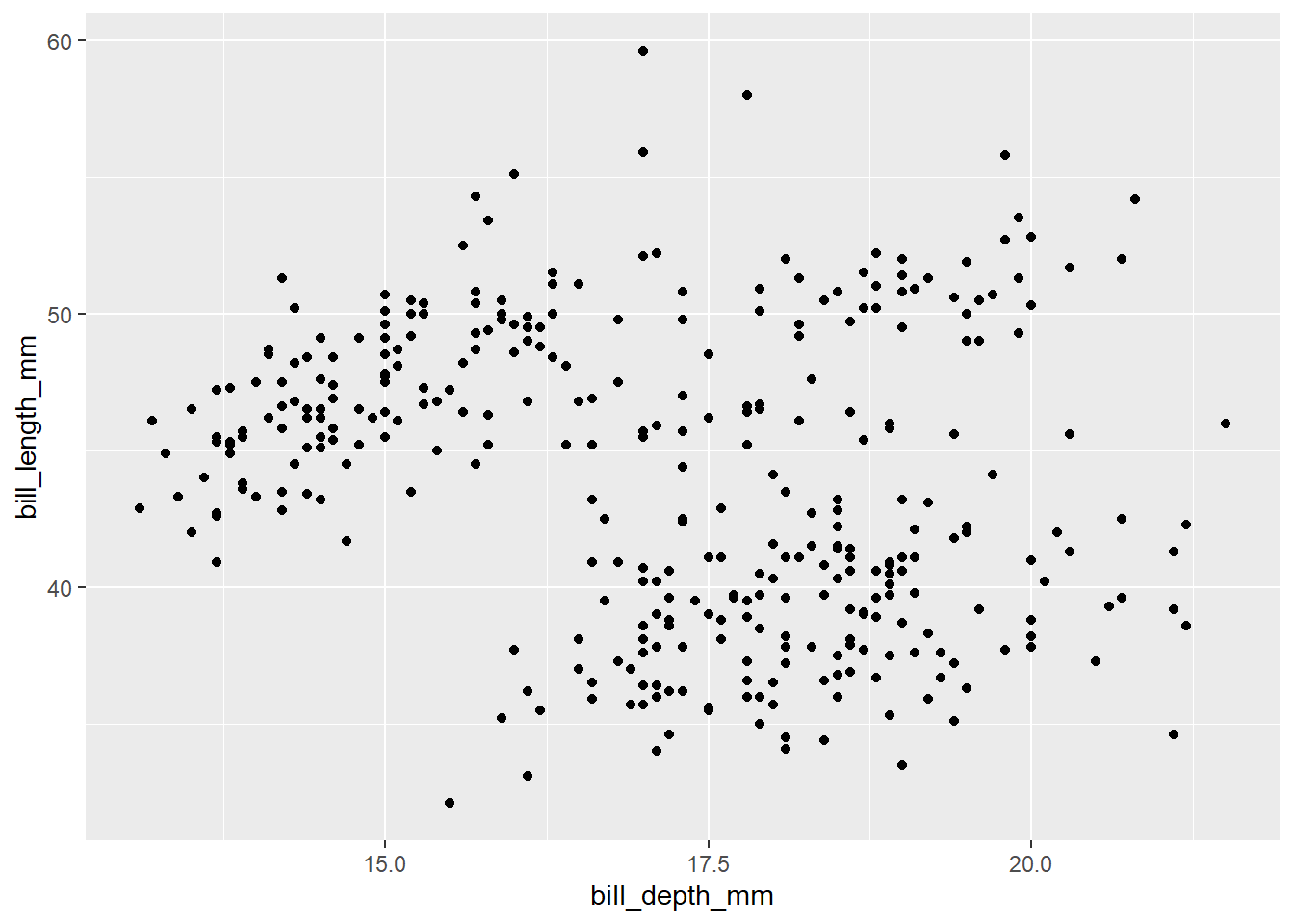
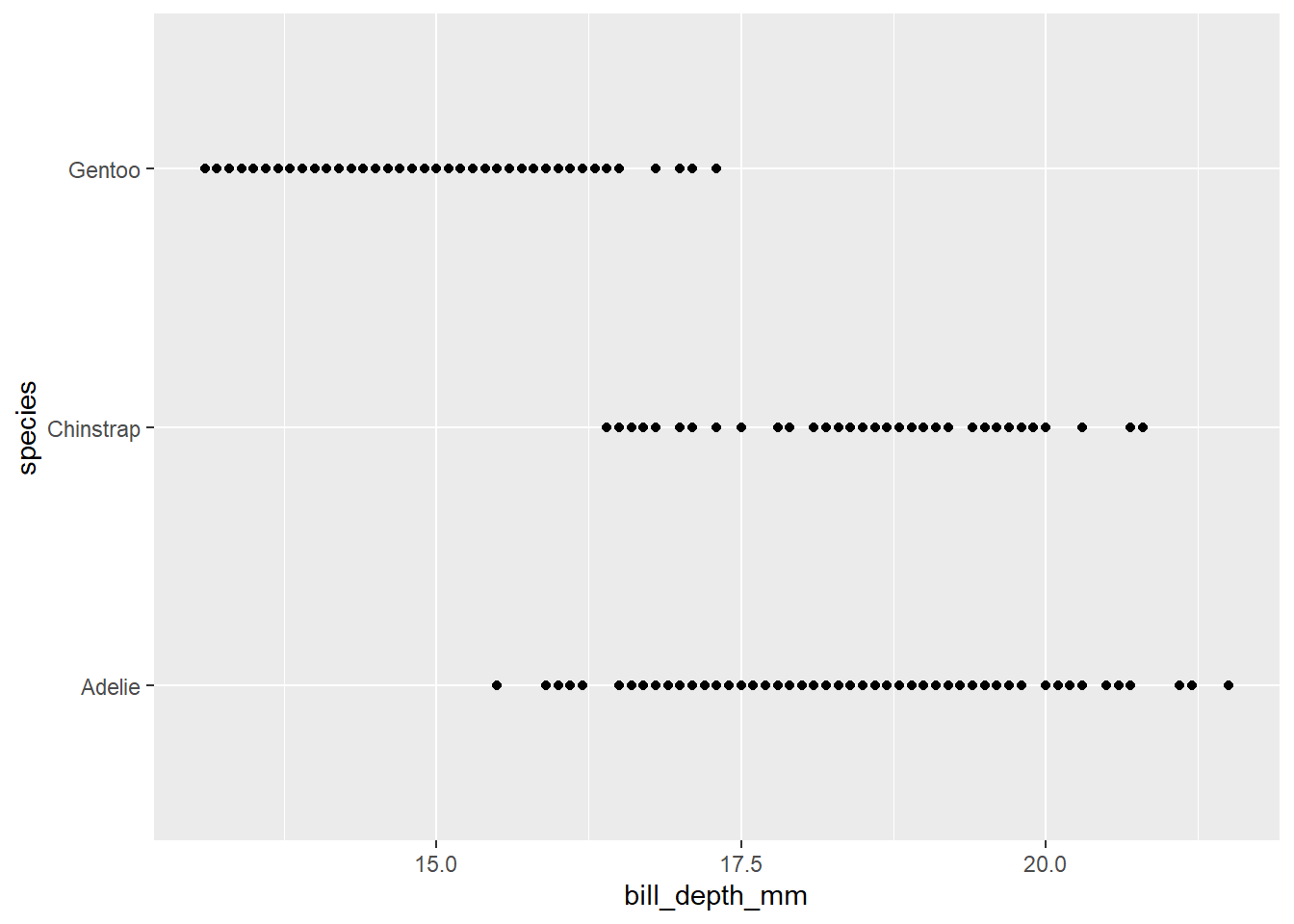
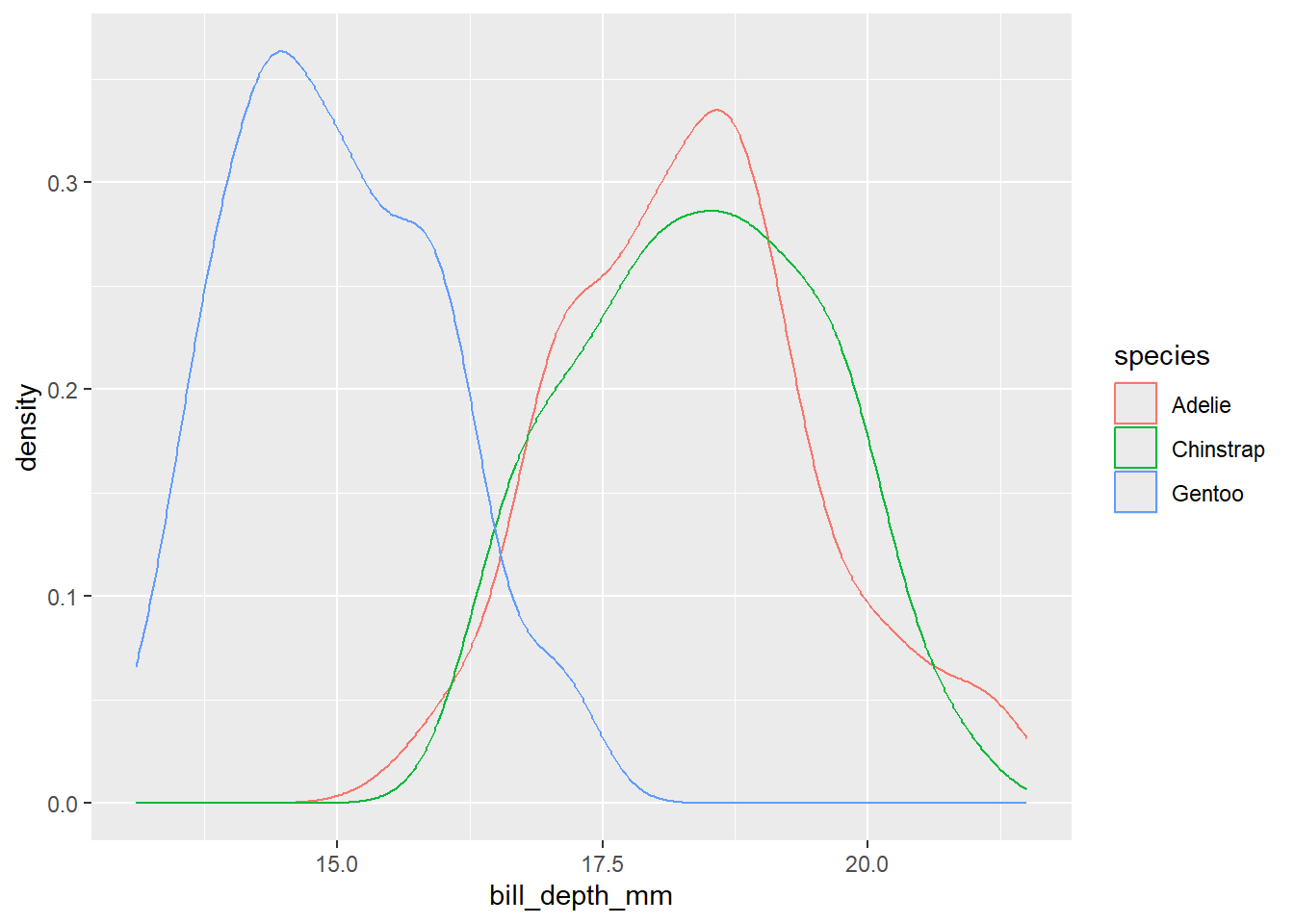
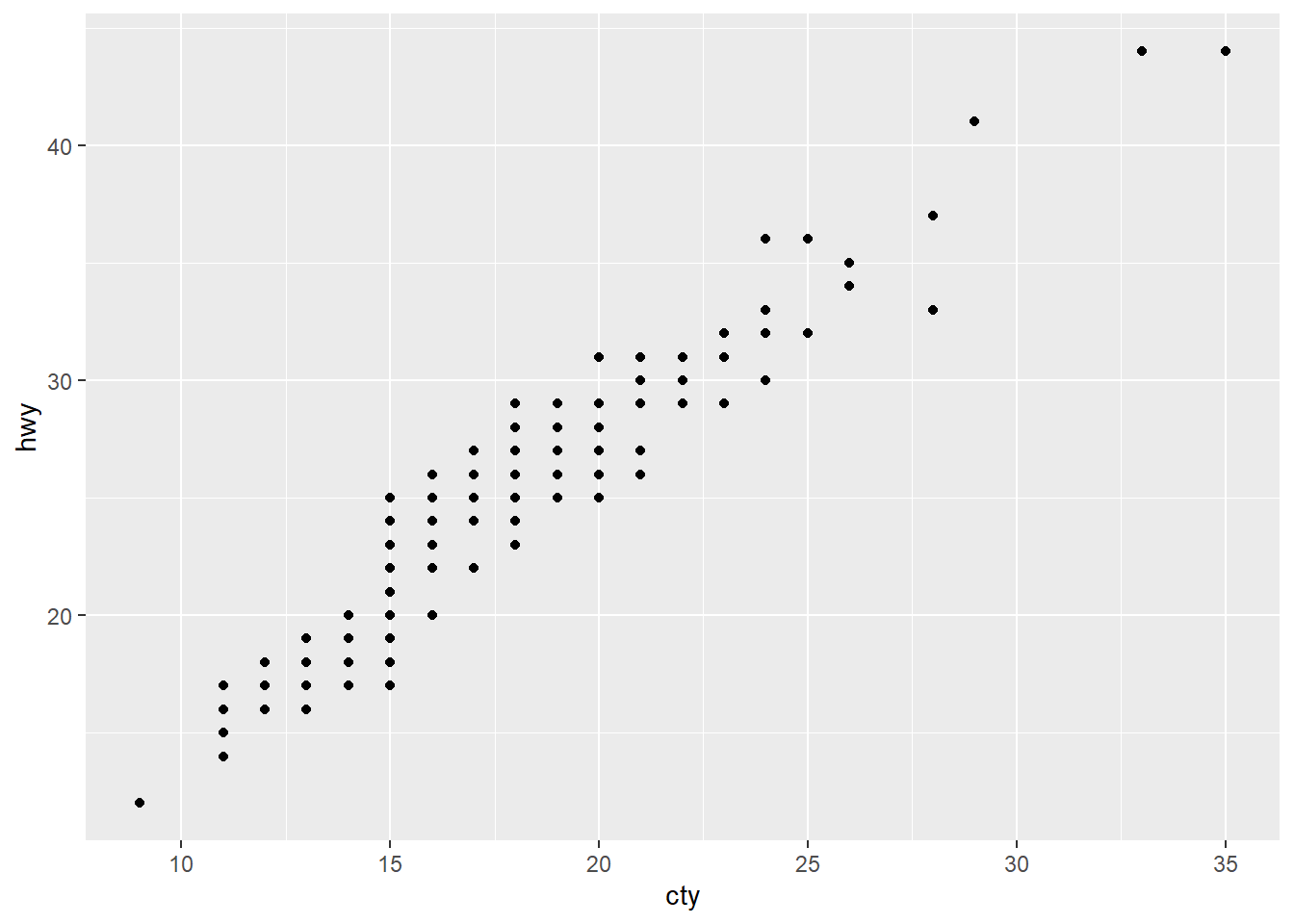
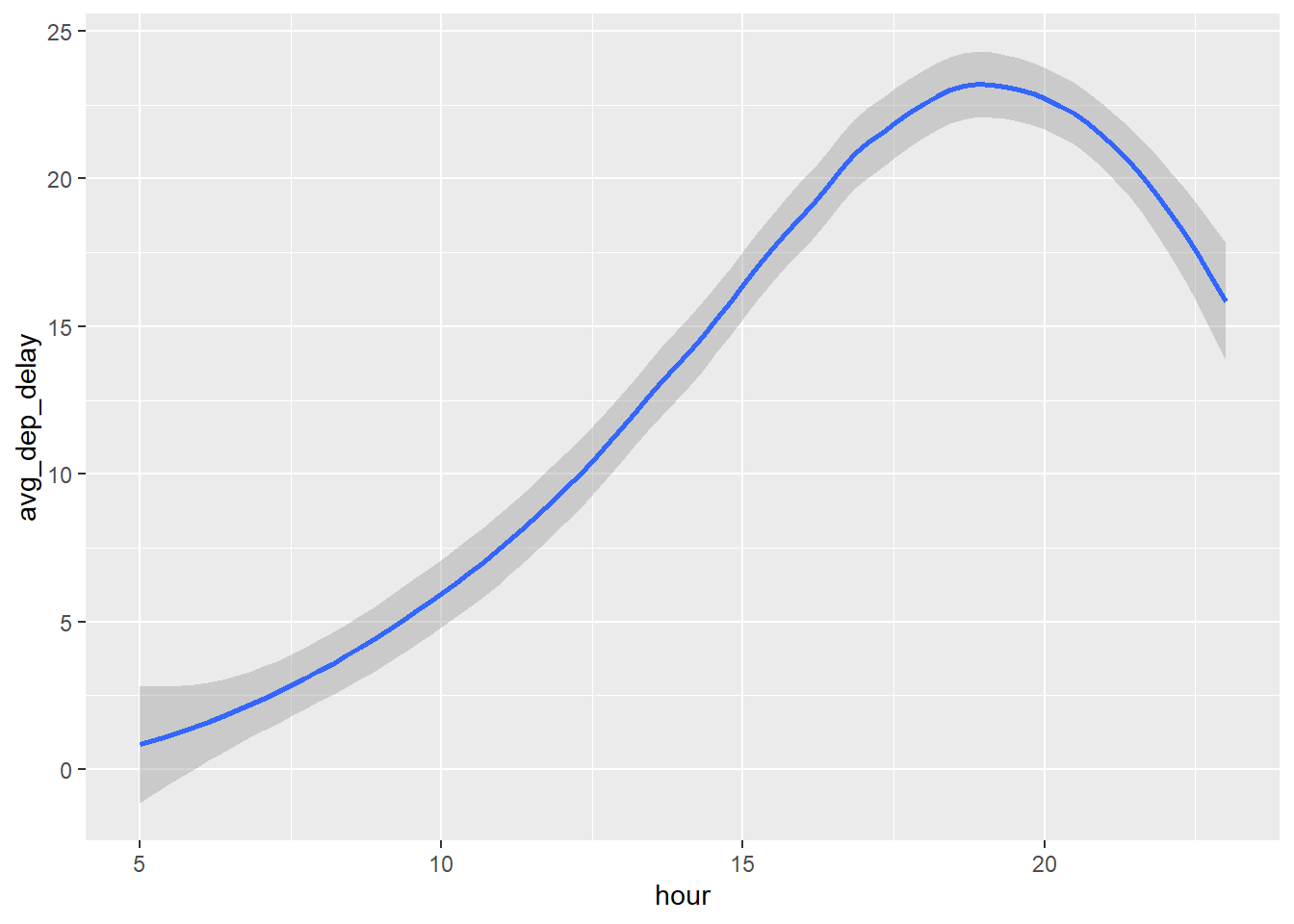
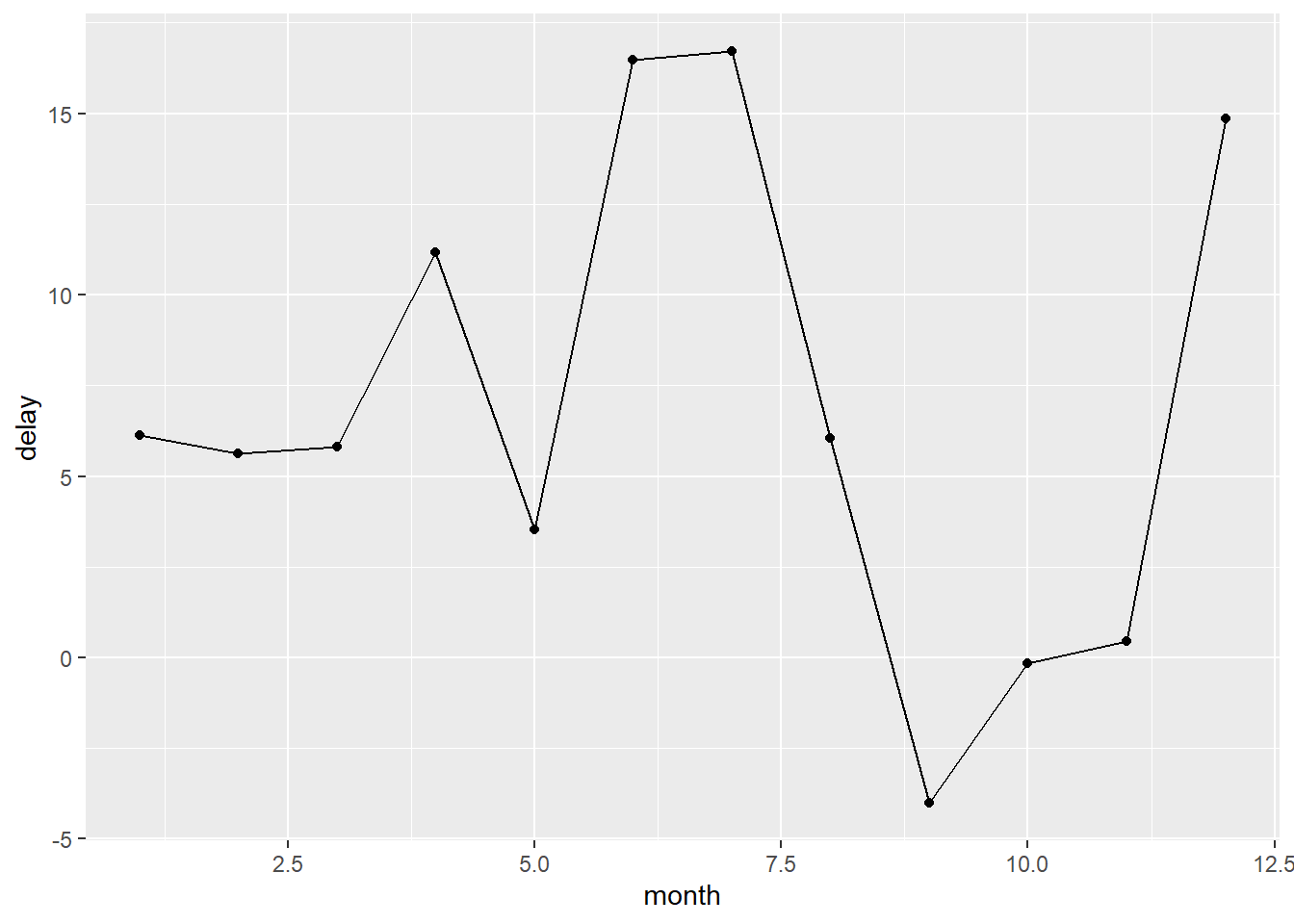
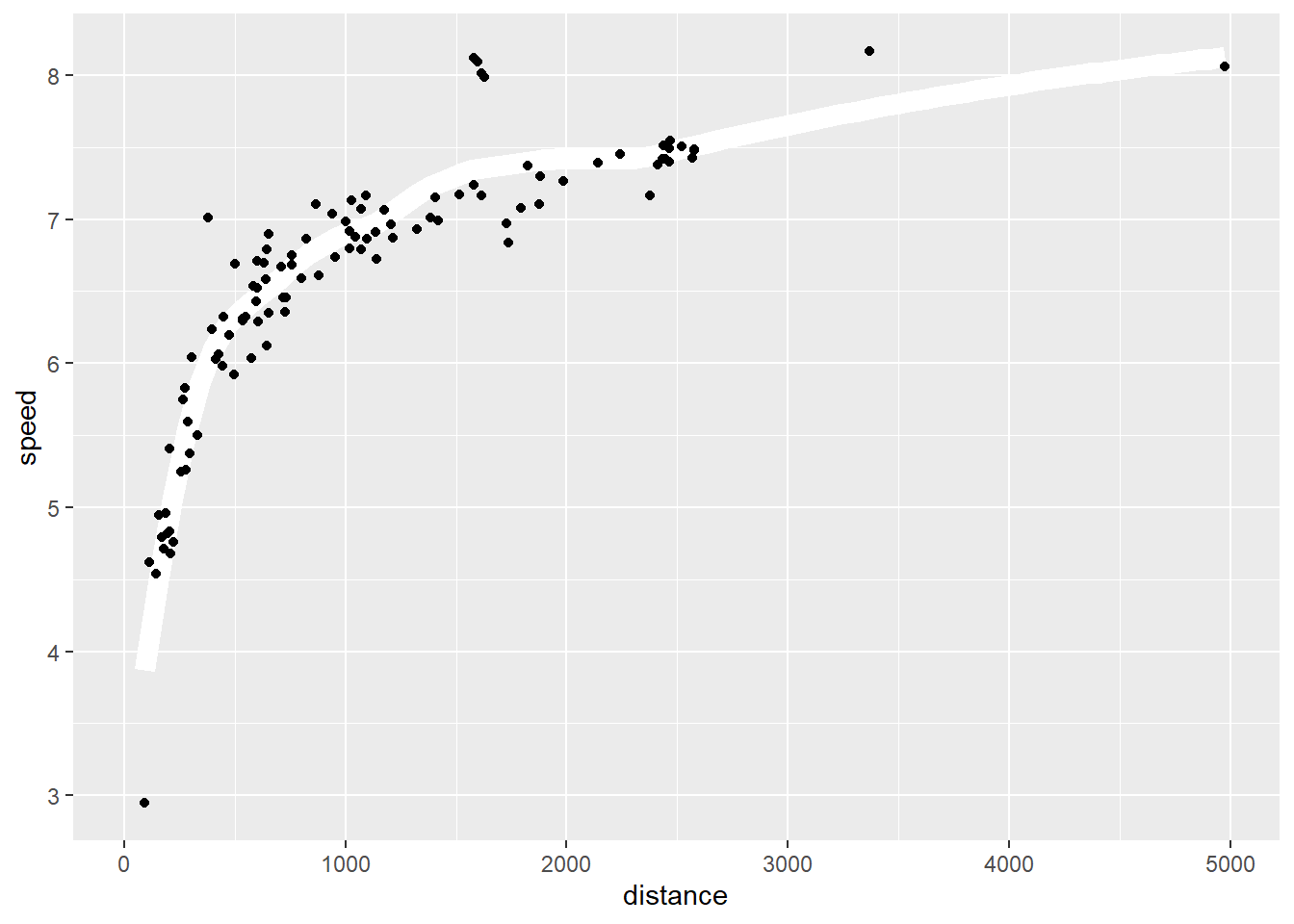
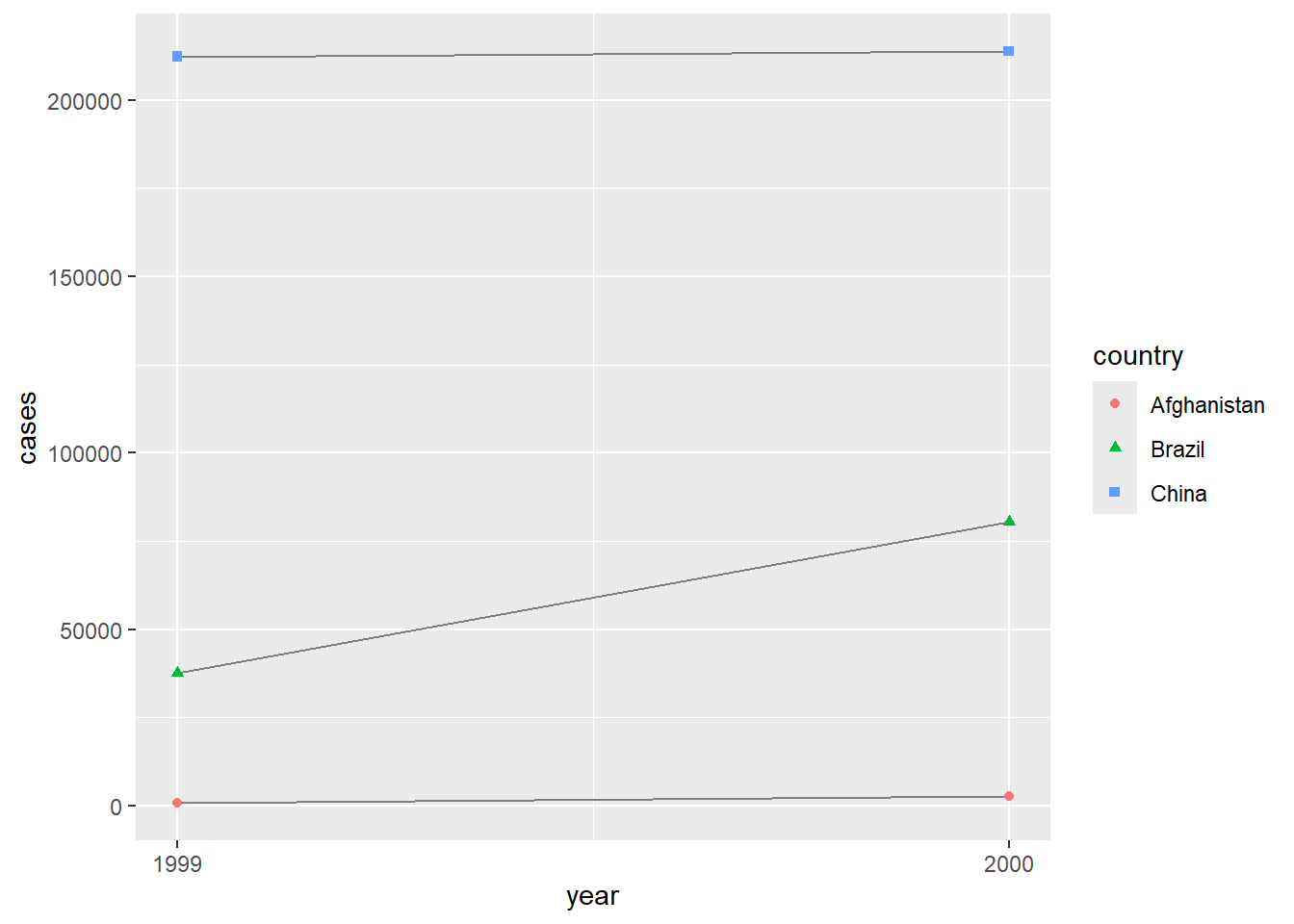
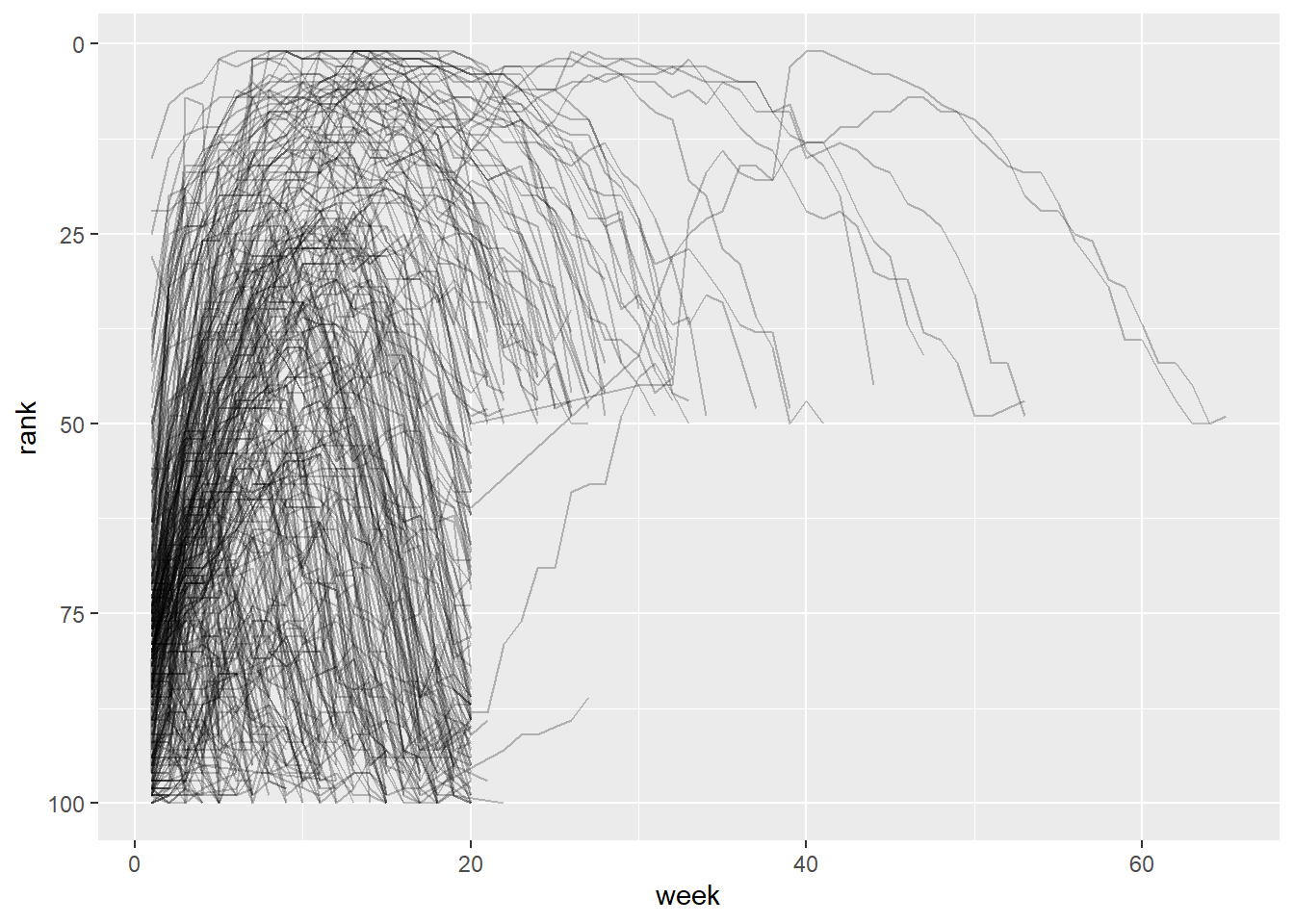
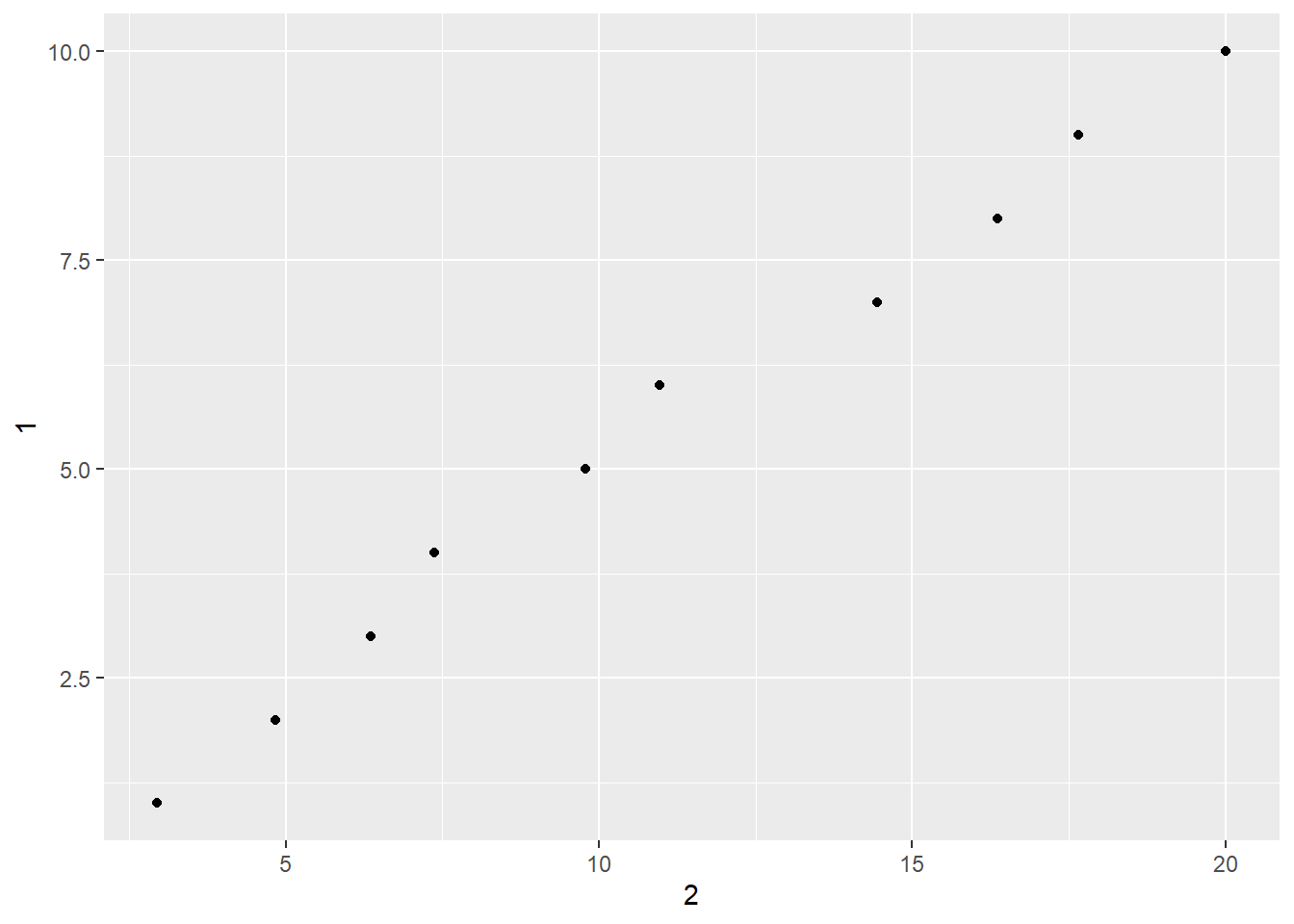
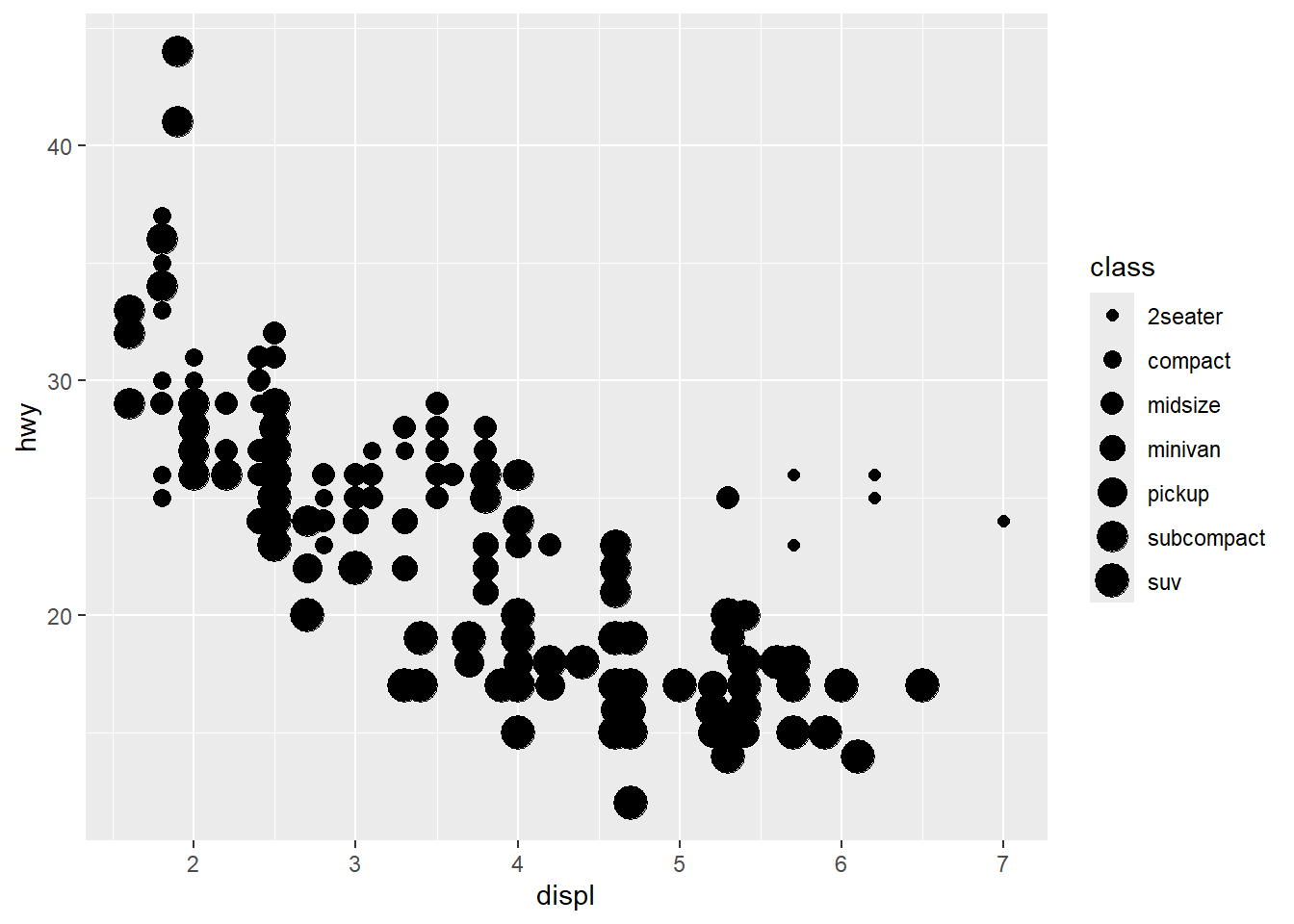
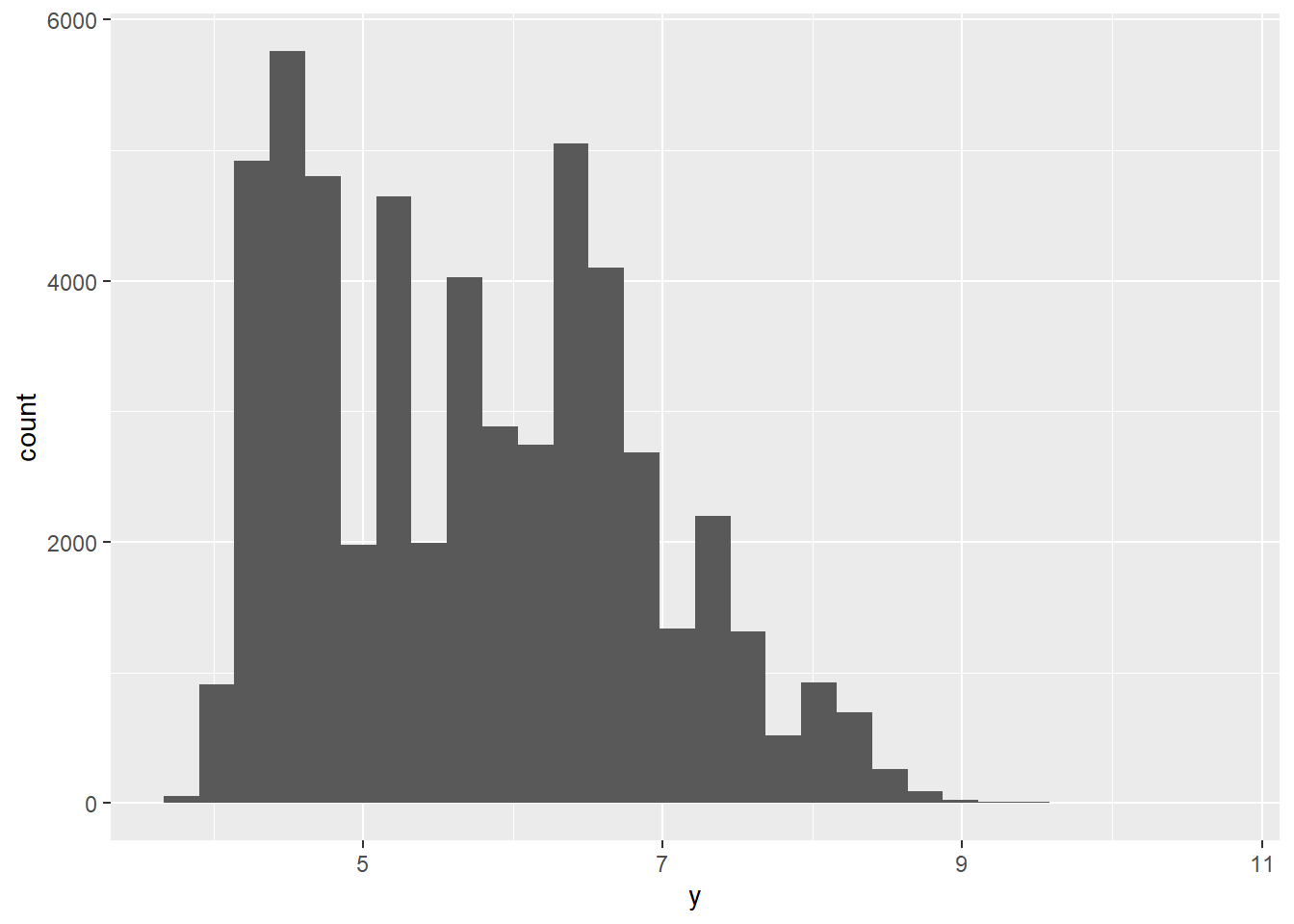
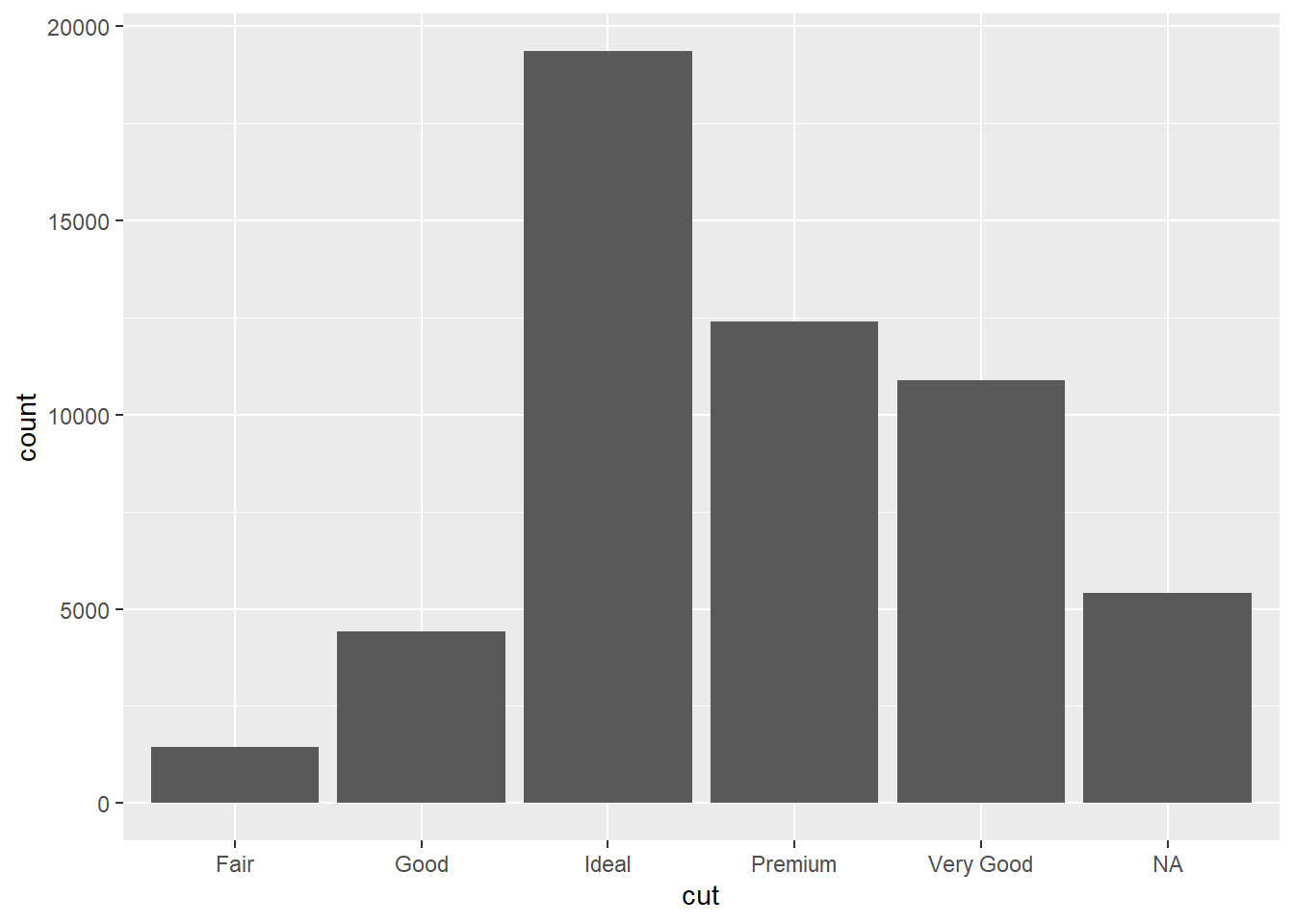
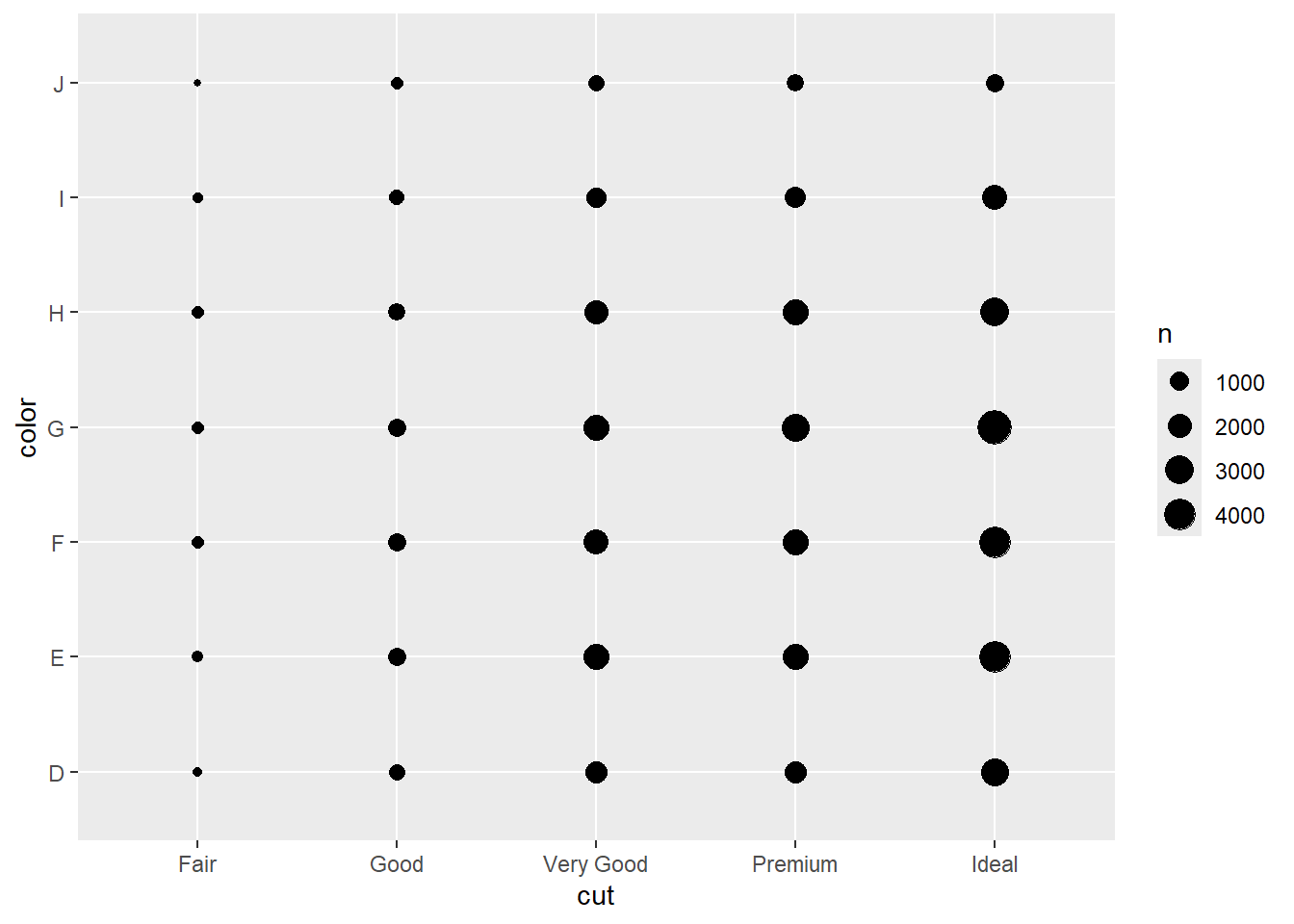
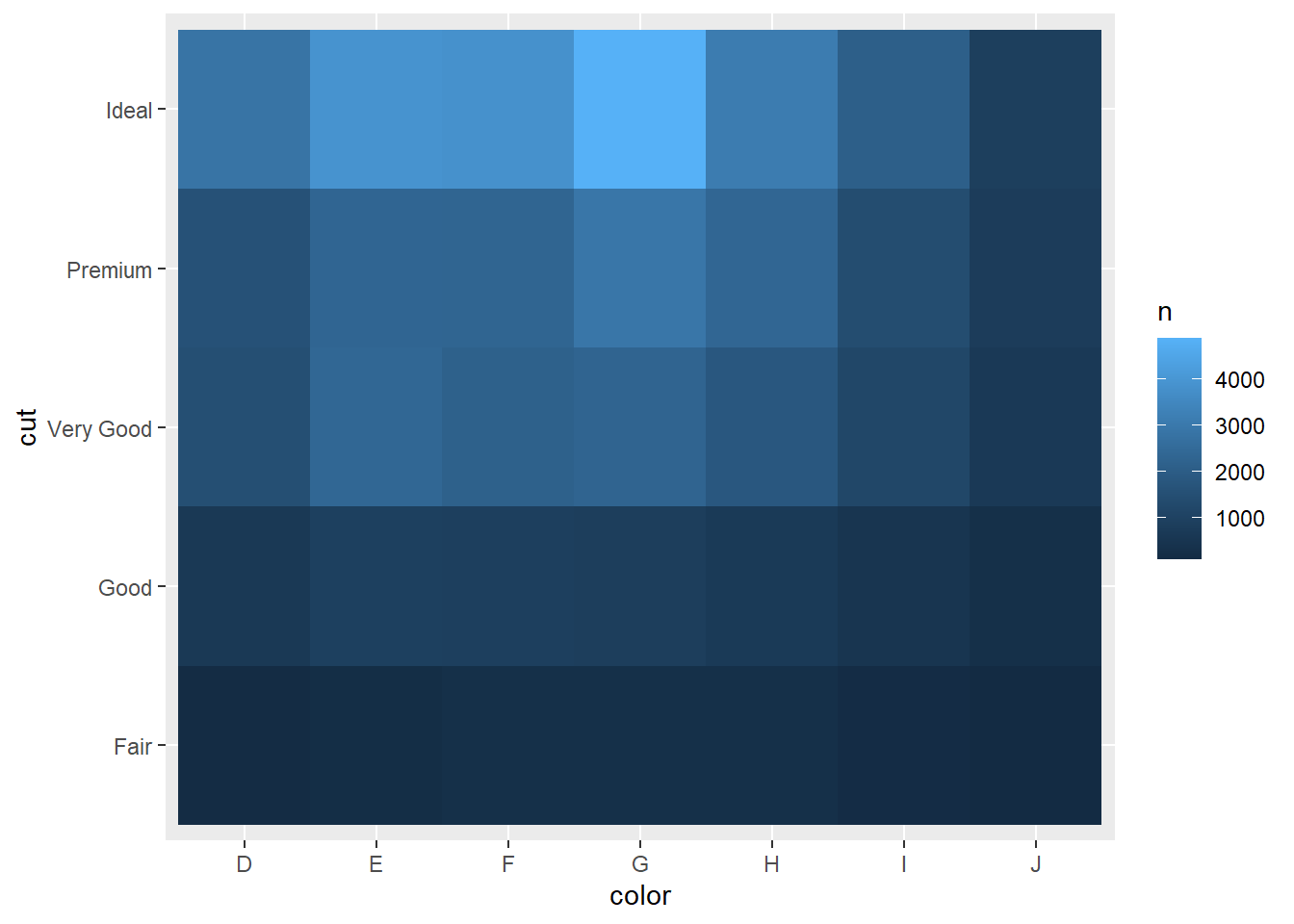
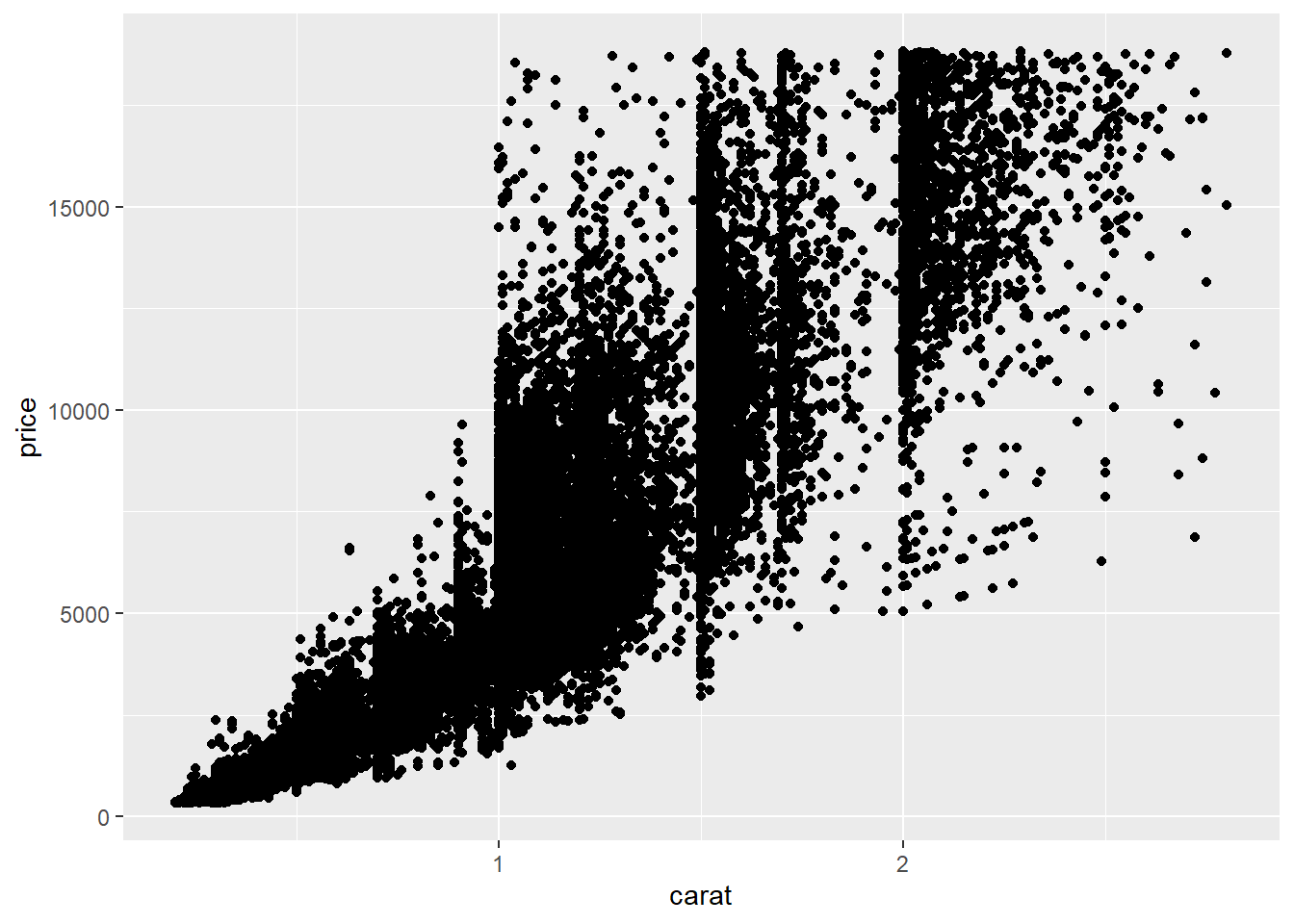
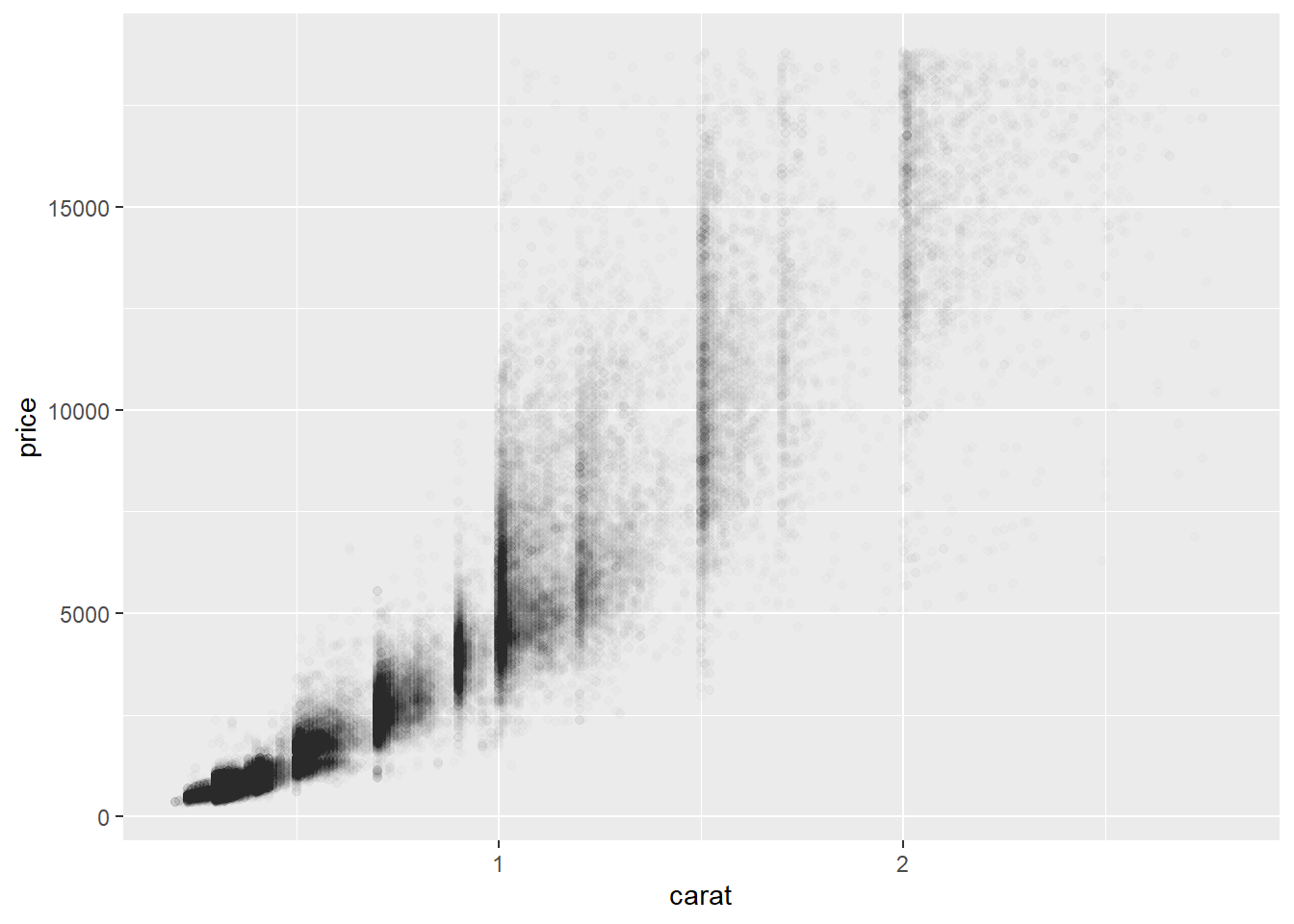
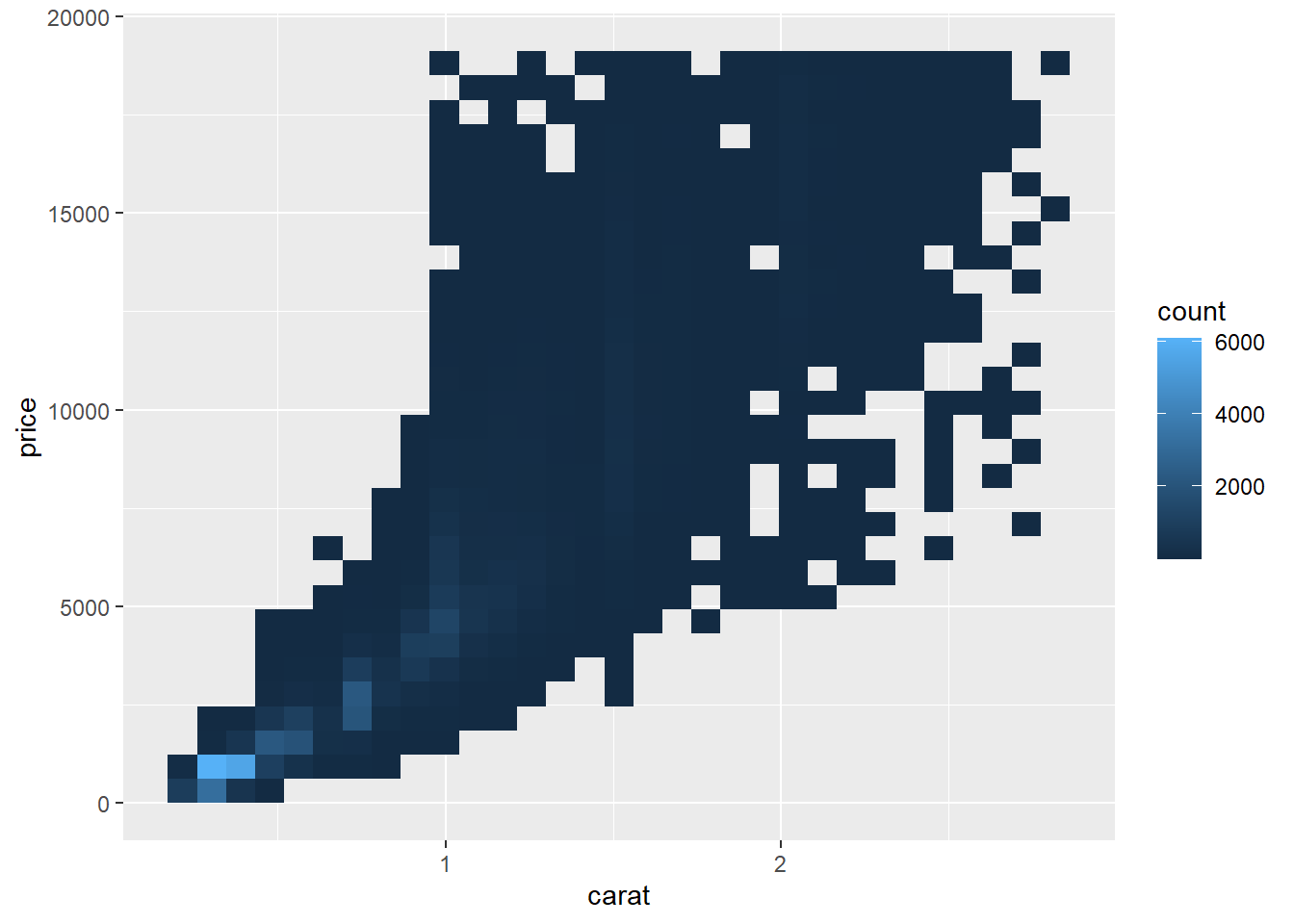

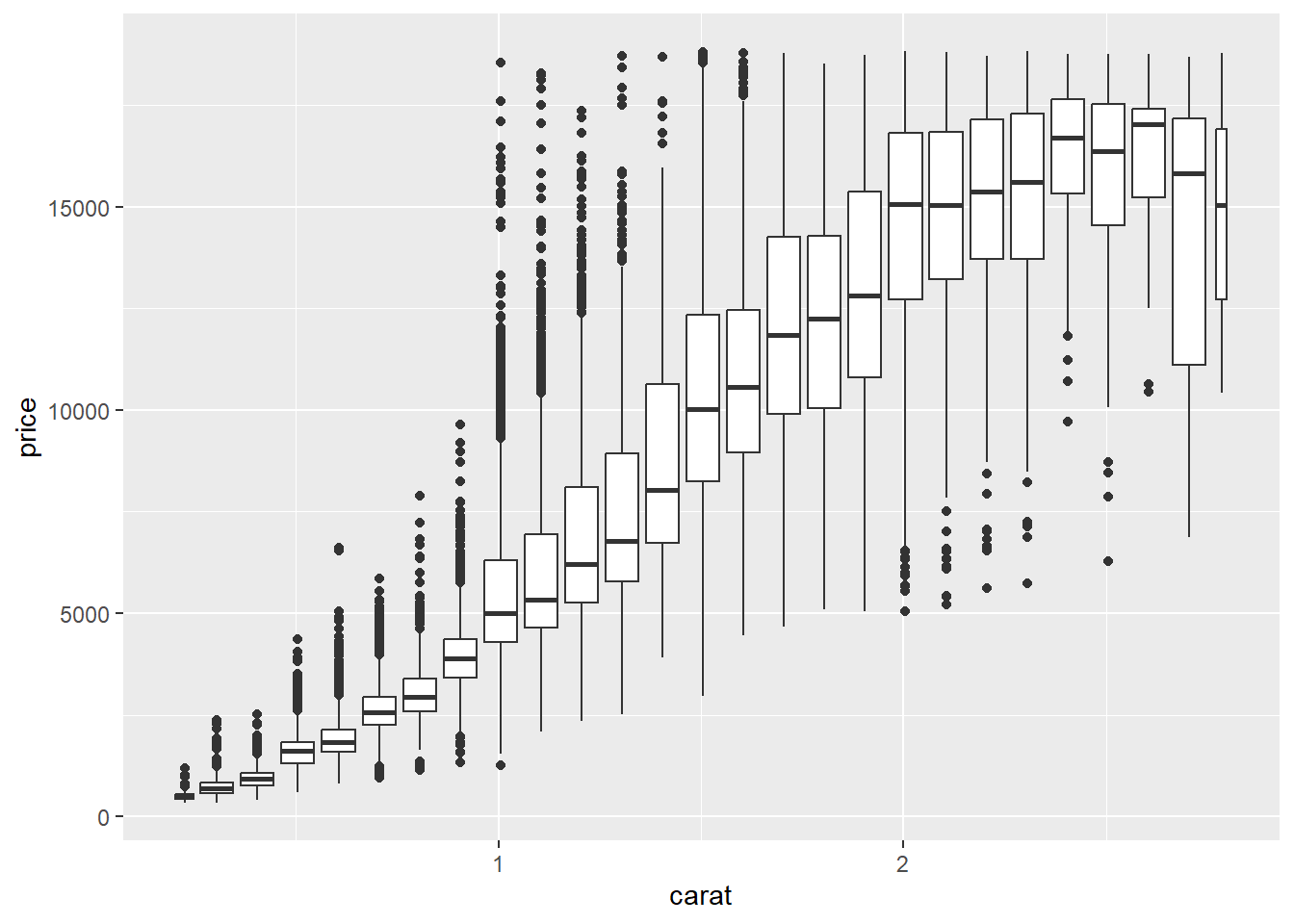
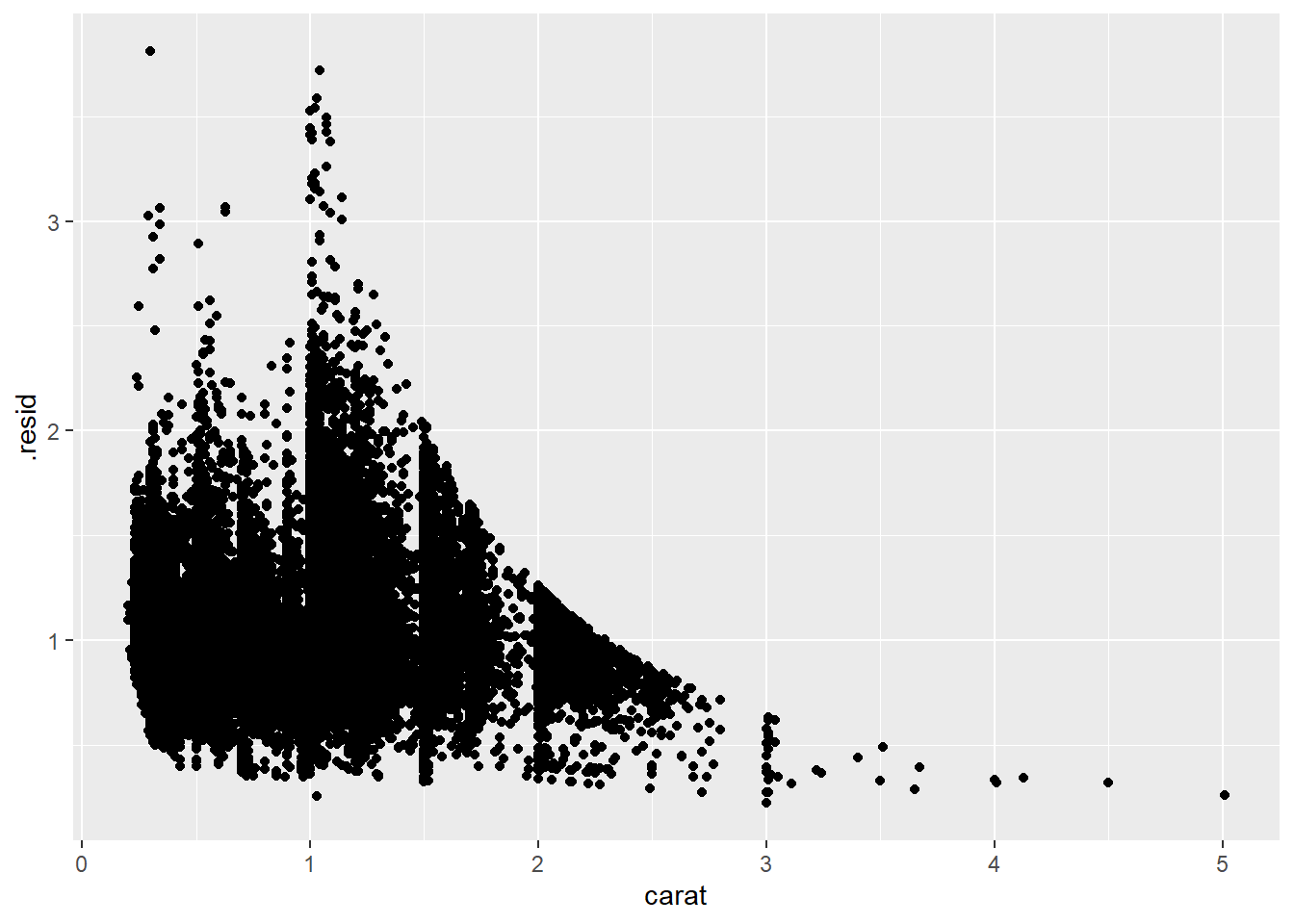
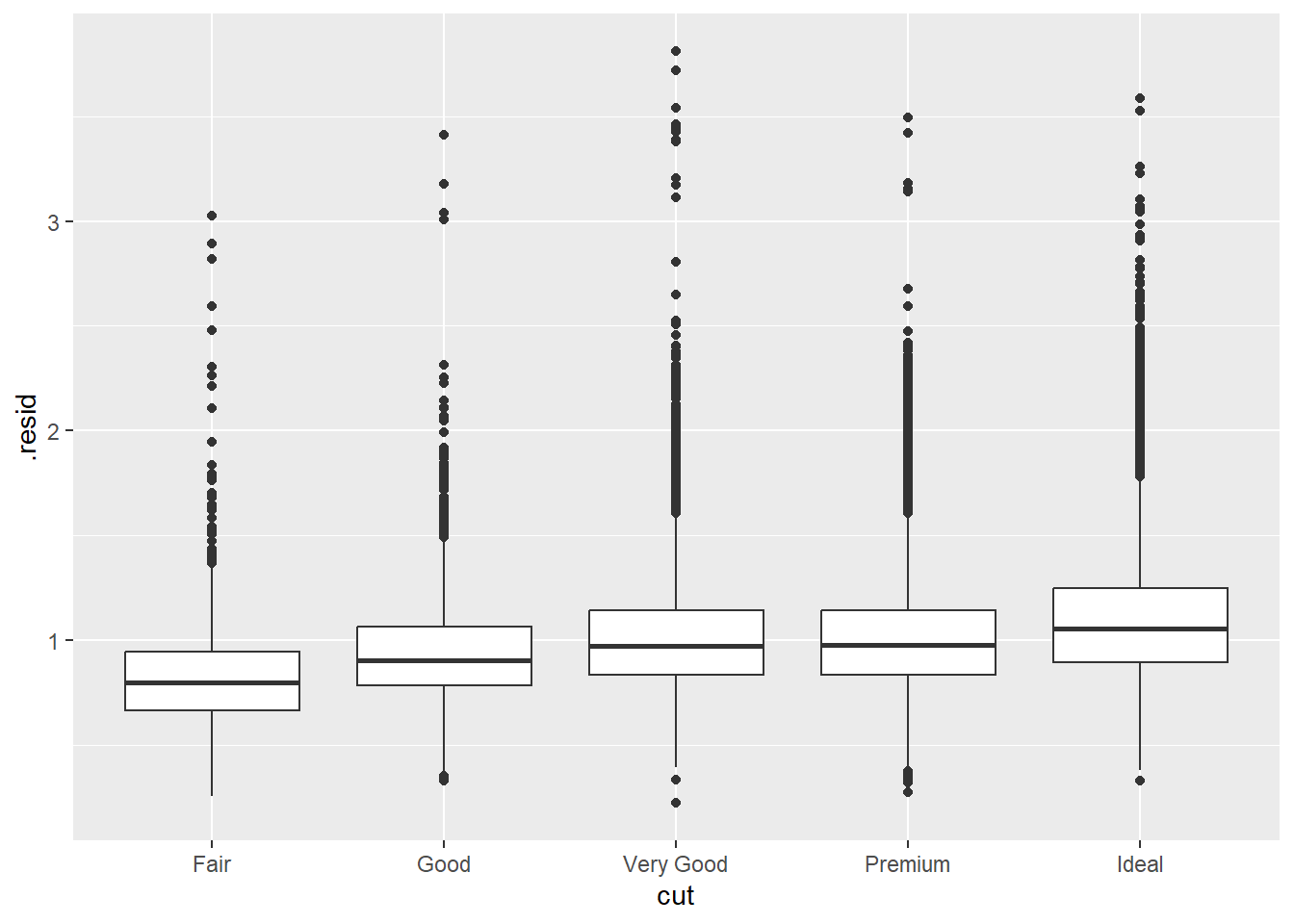
2.2 Comments